Not all cancers are the same, so how can we identify the differences that we can’t see?
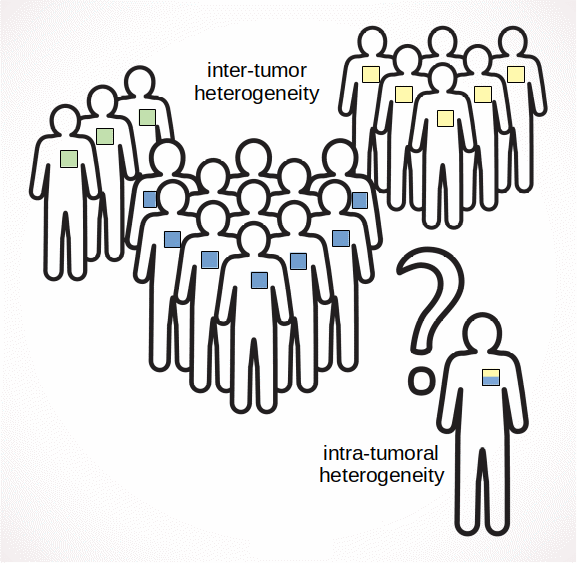
Cancer is not a single disease but a family of diseases with a common characteristic - the uncontrolled division of cells. Even within a single tissue type, not all cancers are the same. Each has a distinctive morphology and genomic properties, responds differently to treatment, and has a unique prognosis. Researchers have found that tumors can be placed into groups, called subtypes, that do have similar properties. Knowing the subtype of a tumor helps doctors in choosing a course of treatment.
Although these subtypes are distinct, some tumors are even more complicated: they contain more than one subtype, called intratumoral heterogeneity. This can cause problems in determining the subtype of a particular tumor. The implications of heterogeneity on prognosis and targeted therapies are not yet well understood.
Heterogeneity Through Images
Subtypes are commonly assigned using molecular properties from a single sample of tumor tissue; this does not provide insight into heterogeneity. A microscopic image of tumor, on the other hand, provides a view of the diverse tissue appearance. Hematoxylin and eosin (H&E) stained histology images are the standard approach for cancer diagnosis by a pathologist. However, identifying the molecular subtype of a tumor from an H&E image is not possible, even for the best trained pathologists. Computers using AI technology have recently learned to assign tumor subtype from histology images, but they also get confused by heterogeneity.
To address this challenge, I created software to better understand the heterogeneity in images. This helps in forming better predictions for the subtype of an individual tumor and also identifies where heterogeneity may exist. The challenge came in that new machine learning methods were needed to solve this problem.
Machine Learning Models
Most machine learning methods train on an input, such as an image, and an output label, such as the subtype. But when multiple images of diverse appearance are provided for a single output, it can get confused. This is called a weakly supervised learning problem.
The subtype labels for training are provided by molecular methods that assign a single subtype from a small piece of tissue, providing no indication of whether a particular tumor is heterogeneous. In working with histology, we take images from different areas of the tumor but still only have a single subtype assigned. Each image from a sample, or each smaller region within a larger image, is referred to as an instance. Following with that terminology, we refer to this as a multiple instance learning problem. Developing a machine learning model to predict the overall subtype of the tissue requires an extra layer of logic: individual instance predictions are combined into a prediction for the tumor.
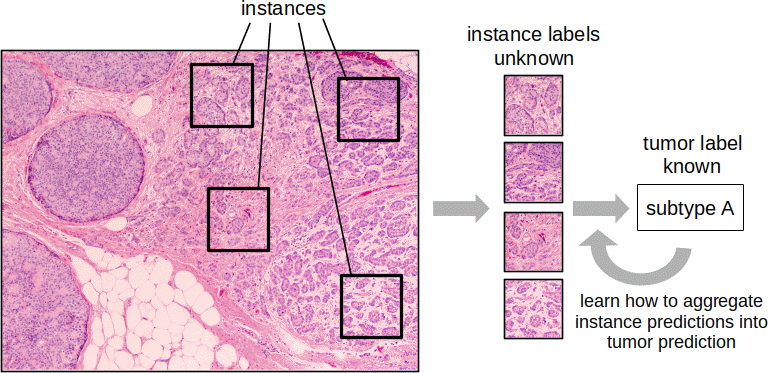
The traditional machine learning approach was to assign the patient subtype to each instance, train a classifier on those instances, and predict for the tumor as a whole with a majority vote over the instances. In the deep learning world, I integrated this idea by adding a multiple instance layer to the network to perform the aggregation, enabling end-to-end learning. The model takes a set of instance images as input, a single patient subtype prediction as output, and learns the features in between to optimize the subtype classification task over a set of training examples. More recent alternatives treat this as an attention problem: learn the relative importance of each instance in predicting the tumor subtype. In other words, they learn not just how to predict the subtype of an instance but also a weight to use in aggregating instances into a tumor prediction.
New Insights
The benefits of a multiple instance model are not just improved subtype predictions for heterogeneous tumors but also some intuition for where that heterogeneity lies. Once the model is trained, it can be applied to novel images and used to predict the subtype for each instance (or local image region) with the results visualized to show which tissue regions are associated with each subtype. The amount of heterogeneity present in each tumor can be quantified over a larger set of patient samples, providing an indication of how much subtype heterogeneity is present overall. Further work is still needed to validate the predicted heterogeneity.
If employed in a clinical setting, identifying the likely presence of intratumoral heterogeneity can influence decisions in obtaining further diagnostics. If heterogeneity is likely, multiple samples from throughout the tumor may be processed to get a more complete assessment. It could also affect treatment decisions as heterogeneous tumors might be more aggressive.
Such powerful insights are due to the large spatial dimension of histology images. Most genomic techniques cannot provide such spatial precision. Until single cell genomics becomes more prevalent, deep learning analysis of large images showing the diverse tissue appearance are our best view into intratumoral heterogeneity.
Would you like help implementing deep learning-based methods to quantify tissue heterogeneity?
I’ve worked with a variety of teams to develop advanced machine learning algorithms to extract new insights from pathology images. Schedule a free Machine Learning Strategy Session to get started.