Portfolio
Power Plant Emissions from Remote Sensing Imagery
Background: This project with WattTime aims to estimate carbon emissions from all large power plants worldwide using satellite imagery. The images come from a variety of satellites, sensors, and wavelengths that observe power plants at different times and capture different properties.
Challenge: We are limited by the revisit times of satellites, low spatial resolution, and partial occlusion from clouds. Our models must also generalize from countries with recorded emissions on which we can train to all power plants globally. Remote sensing images are quite different than everyday photographs. They contain different types of objects and textures, vary in appearance from one satellite to another, and have more than three color channels.
Solution: This project is still ongoing, but our efforts so far have focused on visible signals from power plants: vapor plumes emitted from cooling towers, mechanical cooling, and emissions control technology. We developed deep learning models that focus on the sources of these signals to predict whether a plant is on or off and estimate its current generation. These predictions are aggregated across different models and satellites, as well as over time, to produce emissions estimates for each plant.
Press Release 2019
Forbes
Press Release 2020
IEEE Spectrum
Time Best Inventions of 2020
Climate Change AI Workshop @ NeurIPS 2020
Duke Energy Data Analytics Symposium
Google Sustainability Video
Quartz
COP28: Al Gore and Climate TRACE Unveil Game-Changing Greenhouse Gas Emissions Inventory
Science
YouTube: Al Gore and Gavin McCormick unveil unprecedented Climate TRACE emissions data at COP29
MDPI: Estimating Carbon Dioxide Emissions from Power Plant Water Vapor Plumes Using Satellite Imagery and Machine Learning

Methane Plume Segmentation
Problem: While carbon dioxide is the most prevalent greenhouse gas, methane is much more potent. And methane leaks can be difficult to detect. This project used multispectral satellite imagery to detect and segment methane plumes. The primary challenge in model training was the scarcity of true methane plumes with which to train a model.
Solution: This project involved reviewing data and modeling choices on the project and subsequent steps for refining the training data and model approach in order to advance the project as quickly as possible.
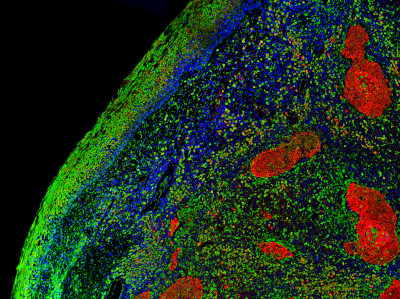
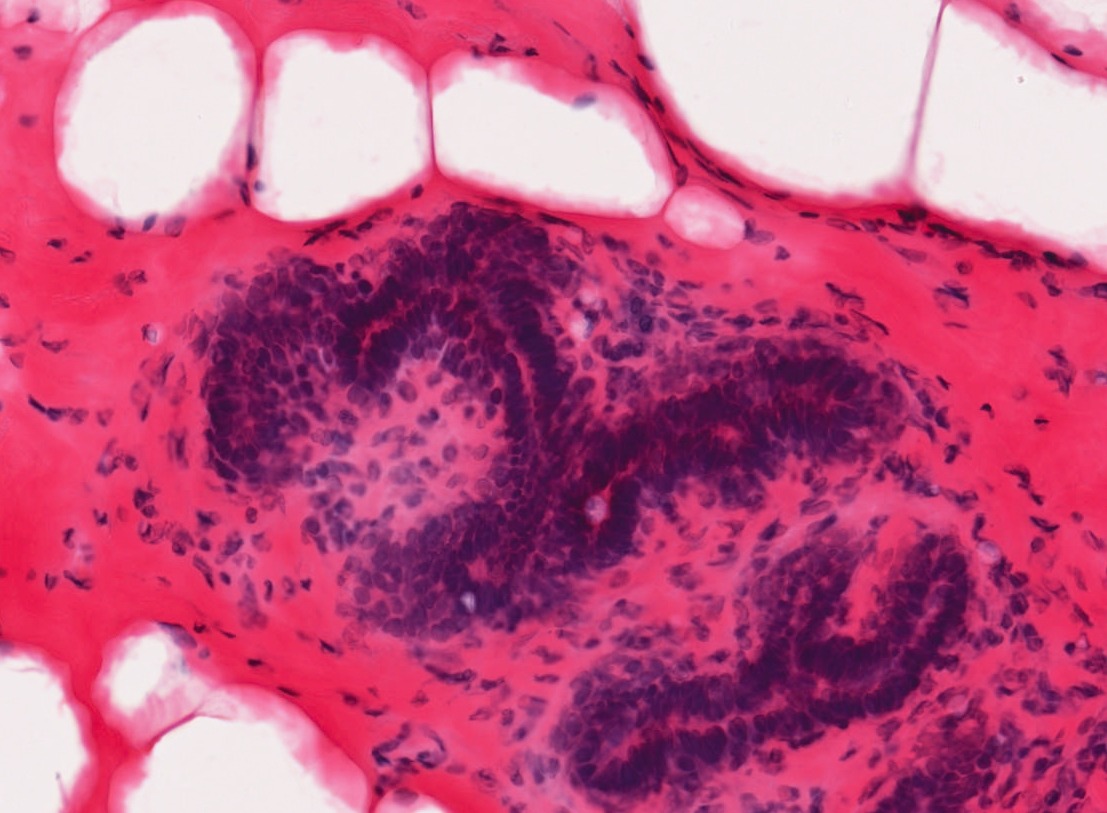
Lesion Classification and Segmentation from Multispectral Images
Problem: H&E histology is the tried and true method for diagnosing cancer and differentiating different types. But an increasing number of alternative modalities are being developed to image tissue. Some don’t require staining, some don’t need slides, and some can even be done in vivo.
Solution: This project involved guiding the development of a tissue classification model using a pathologist’s assessment of H&E as ground truth. The ground truth labels enabled training a weakly supervised model. The multispectral perspective can provide additional information but made transfer learning more challenging. A variety of ways for integrating the different channels were evaluated. Regardless of the machine learning approach, good study design was an essential starting point to ensure that all models developed will generalize to lesions from different patients.
Predictive Biomarkers from Multiplex Immunofluorescence
Problem: While H&E is the most common modality for studying tissue, it only highlights two things: nuclei and cytoplasm. Multiplex immunofluorescence (mIF), on the other hand, characterizes many different proteins and cell types simultaneously. This more complete picture of tissue biology can enable more powerful biomarkers for predicting patient outcomes. However, these additional channels can bring new challenges to training a deep learning model.
Solution: This project began with providing guidance on nuclei segmentation approaches for multiplex imaging. Once individual cell types could be identified, we focused on image translation models that could predict expression levels from a subset of channels. These were used to predict cell types and assess how well different cell types could be predicted.

Tumor Segmentation
Problem: Analyzing gigapixel whole slide images is time-consuming for pathologists, though absolutely critical to the diagnosis of cancer. If cancerous regions can be identified faster for review by a pathologist, the overall analysis can be completed more efficiently. Locating cancerous regions is complicated by the varying appearance of different grades.
Solution: This project involved guiding a junior ML team on best practices for segmenting tissue types. We initially found a number of inconsistencies in the pixel-level annotations created by pathologists, so we started by refining and standardizing the annotation process. As the labeled dataset became cleaner, we were able to iterate more effectively on the segmentation model.

Biomarker Prediction for Precision Medicine
Problem: An overwhelming amount of research over the last few years has demonstrated that a number of molecular tumor biomarkers can be predicted from H&E whole slide images. This is a weakly supervised classification problem as the model must learn which regions of tissue are associated with each class. A number of multiple instance learning approaches have been proposed to tackle this.
Solution: This project involved identifying the most promising approaches for building a robust and generalizable model for predicting a particular molecular biomarker. Open source code bases were used where suitable.
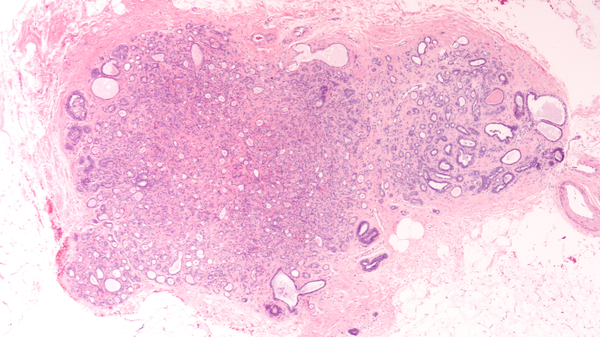
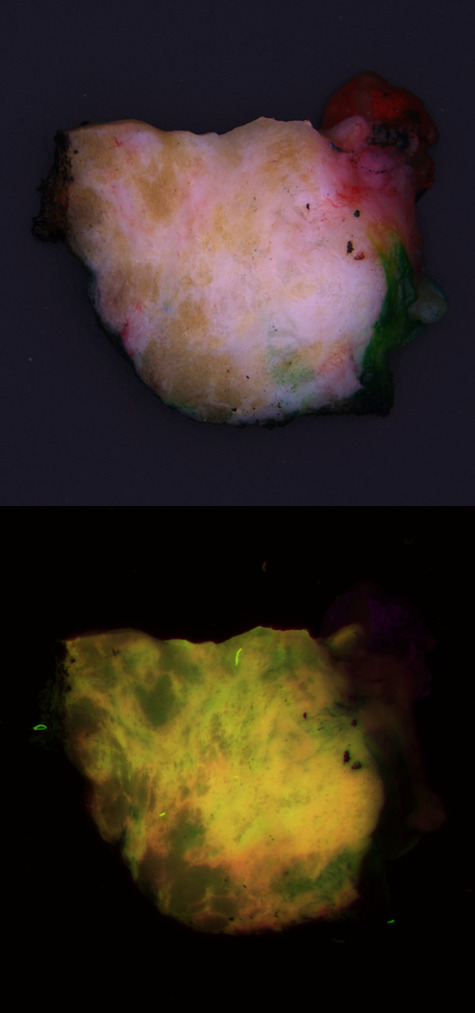
Cancer Detection in Autofluorescence Imagery During Surgery
Problem: Surgeons try to conserve as much healthy tissue as possible, while excising all tumor tissue. However, distinguishing between the two during surgery is challenging. For a definitive call, the tissue must be processed for histological analysis. CytoVeris is developing an ultraviolet-excited autofluorescence platform to image excised tissue during surgery and highlight any regions that are likely to be cancerous.
Solution: I assisted CytoVeris in training machine learning models to distinguish cancer from healthy breast tissue in multispectral autofluorescence images. Pathologist annotations were obtained from the corresponding H&E image and were spatially referenced with autofluorescence images of the tissue. Some of the technical challenges included the diversity and mixed heterogeneity of tissue samples, and the precision of the image to H&E registration, both of which can result in some degree of label uncertainty. We compared a variety of different methods for feature extraction and classification. The model results also provided feedback for data cleaning and model improvement efforts. As lighting and optical techniques are improved in new iterations of the imaging device, the models continually became more robust.
OncoSIGHT AI
Archives of Pathology & Laboratory Medicine: AI-Powered Biomolecular-Specific and Label-Free Multispectral Imaging Rapidly Detects Malignant Neoplasm in Surgically Excised Breast Tissue Specimens
Press Release
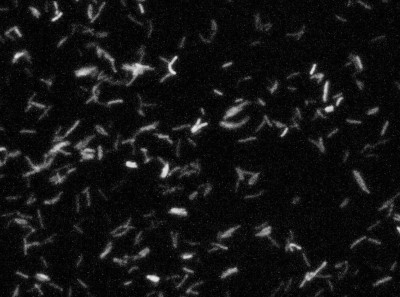
Detect, Segment, and Classify Bacteria
Problem: Counting the number of each species of bacteria is important for a variety of applications, but it is complicated by their small size, overlapping cells, and similar appearance of different species. This task has previously been tackled with traditional image processing techniques to locate objects, estimate cell boundaries, and differentiate species based on cell size.
Solution: Deep learning provides the potential to learn a more accurate method to detect and classify each cell or cell-look-alike. While segmentation may not be strictly necessary, detecting tiny objects is quite challenging and can be more accurate when approached with a segmentation model. Instance segmentation was selected to better handle adjacent and overlapping cells.
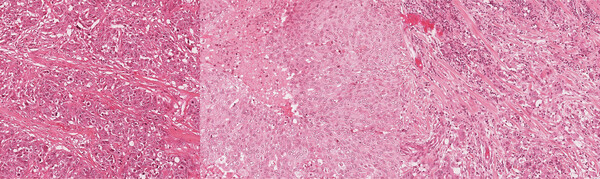
H&E Stain Normalization
Problem: One of the largest challenges in histopathology image analysis is creating models that can handle the variations across different labs and imaging systems. These variations can be caused by different color responses of slide scanners, raw materials, manufacturing techniques, and protocols for staining. Different setups can produce images with different stain intensities or other changes, creating a domain shift between the source data that the model was trained on and the target data on which a deployed solution would need to operate. When the domain shift is too large, a model trained on one type of data will fail on another, often in unpredictable ways. The goal of stain normalization is to standardize the appearance of these stains.
Solution: The goal of this project was to evaluate a variety of stain normalization methods and their effectiveness in reducing domain shift for downstream analysis tasks. We ran both traditional and deep learning-based methods on three different H&E datasets that include whole slide images from different scanners. We assessed each with respect to accuracy on a downstream task, ease of training, computation requirements, and failure modes.
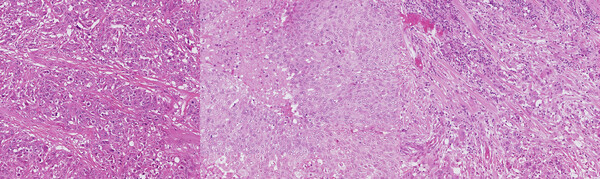
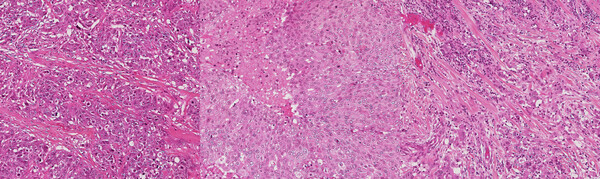
Improving the Generalizability of Histopathology Classification
Problem: Training models that are robust to staining and scanner variations is one of the largest challenges in computational pathology. The most common solutions include color augmentation, stain normalization, and domain adversarial training. Model generalizability can be further impeded by variations in the patient population.
Solution: This project involved digging deep to reveal as many tactics as possible for improving model generalizability across a large set of models for different cancers and biomarkers. Solutions included histopathology-specific variants of color augmentation and domain adversarial training, as well as pretraining approaches like self-supervised learning. By adapting methods to the unique characteristics of H&E whole slide images, we can build models that generalize better to the variations present in these images.
Whole Slide Image Quality Control
Problem: Whole slide images always come with some processing artifacts: tissue folds, marker lines, uneven sectioning, bubbles, out-of-focus regions, etc. However, machine learning models typically don’t handle these detects in a robust way – especially if they were excluded from the training data. Therefore, image quality control is essential.
Solution: This project involved researching and reviewing published articles on detecting a variety of artifacts, followed by formulating a plan for detecting the most common defects. Some can be detected by fairly simplistic algorithms, but deep learning is usually the most accurate – given sufficient training data. Some artifacts can be simulated to create additional training data, while others require extensive annotation efforts.
H&E Image Analysis
Problem: A variety of deep learning methods have been applied to H&E whole slide images for quality control, anomaly detection, stain normalization, nuclei detection and segmentation, and tissue segmentation. But which approaches are worth trying?
Solution: This project involved researching a variety of image analysis tasks for H&E whole slide images to identify those that are most successful but also easy to implement or have open source code available. These approaches were used to rapidly advance multiple projects.
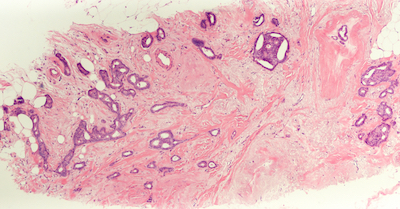
Mitosis Detection in H&E
Problem: One of the hallmarks of cancer is rapidly dividing cells. Histopathology can catch a snapshot in time, with the stages of cell division (mitosis) visible. Pathologists count mitotic figures to assess the aggressiveness of a tumor.
Solution: This project involved reviewing research on mitosis detection to identify the most promising approaches. Object detectors based on YOLO are extremely fast but have low recall on small objects, while ones based on Faster R-CNN perform better on tiny objects like mitoses. RetinaNet can provide further improvements by focusing the loss on hard examples. For fine-grained classification tasks, a second stage classifier may even be necessary to discard more challenging mitosis look-alikes.
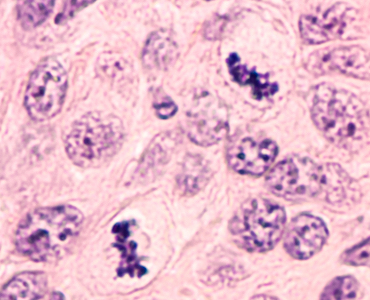
Multiplex Tissue Analysis
Problem: While H&E is the most common modality for studying tissue samples, an increasing number of multiplex options are becoming available. Multiplexing enables the study of multiple markers simultaneously. However, deep learning on multiplex imagery has not been studied nearly as much as H&E.
Solution: This project involved developing a strategy for applying deep learning to multiplex images of cancer and involved research into segmenting and classifying cells and tissue.
Tumor Subtype Prediction with Convolutional Neural Networks
Problem: A growing amount of research has demonstrated the ability to predict tumor genomic subtypes from H&E histopathology. But this capability may be stronger for some cancer types and biomarkers than for others. This project evaluated the ability of a baseline convolutional neural net solution to predict genomic subtype for a cancer type that had not been previously evaluated in this manner.
Solution: Using open source research code and transfer learning, this project measured the classification accuracy of a multiple instance learning model to predict genomic subtype.
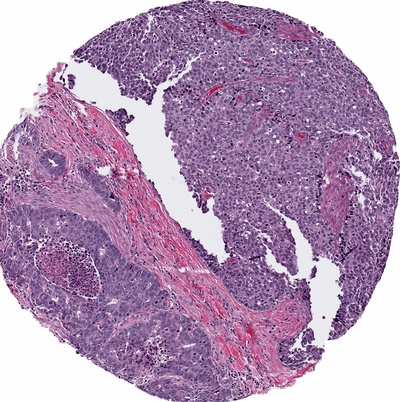
Image Analysis with Deep Learning to Predict Breast Cancer Subtypes
Background: Molecular tests are widely used for patients with some types of breast cancers; however, they can be costly and are not available in many countries. Methods for inferring molecular subtype from histologic images may identify patients most likely to benefit from further genomic testing.
Challenge: Some tumor properties like grade are assigned by a pathologist from histology. Others are computed from molecular properties and were not previously known to be predictable from images. Our methods were the first to test whether estrogen receptor status and genomic subtype can be predicted from H&E histology.
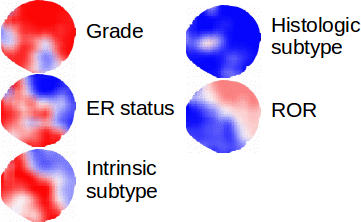
Solution: We developed an image analysis approach using deep transfer learning. A training set of breast tumors was used to create image-based classifiers for tumor grade, ER status, PAM50 intrinsic subtype, histologic subtype, and risk of recurrence score. The resulting classifiers were then applied to an independent test set.
Results: Histologic image analysis with deep learning distinguished each of these classes with high accuracy. Our method was the first to demonstrate that molecular and genomic properties of breast tumors can be predicted from H&E histology. We also demonstrated a visualization technique to show which parts of the tumor are associated with each class.
Additional Applications: Given sufficient labeled data, computers can learn concepts much more complex than even the best trained human experts. While molecular subtypes of breast cancer is one example, machine learning, and deep learning in particular, can provide suitable methods for many complex tasks from assessing prognosis of cancer to predicting earthquake aftershocks. The key is a sufficiently large set of labeled data - the more the better.
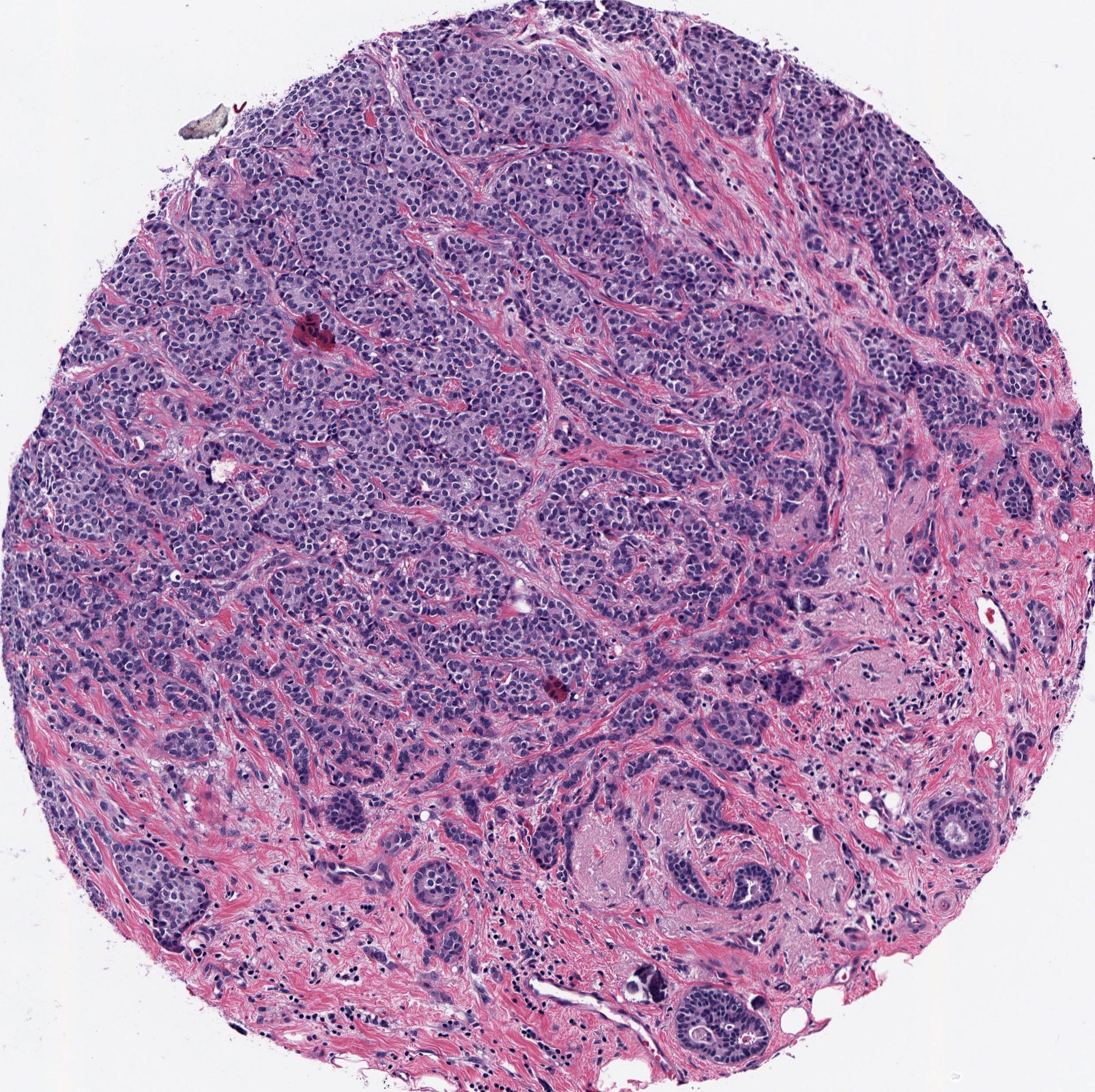
Multiple Instance Learning for Heterogeneous Images: Training a CNN for Histopathology
Background: Genomic subtypes for tumors are assigned from a single sample of tissue; they do not provide a spatial view of the tumor. Although some tumors are mostly homogeneous, others are very heterogeneous - both in appearance and molecular composition. Histology enables us to examine the spatial heterogeneity of the tumor in a way that gene expression cannot.
Challenge: Breaking large images into small regions for making class predictions is the first step in accommodating heterogeneous images. While prior work has explored different ways to aggregate predictions into a single image classification for some problem types, more general aggregation approaches are needed for heterogeneous images. Further, instance aggregation techniques in a deep learning setup can help adapt to this weakly labeled image scenario.
Solution: Multiple instance (MI) learning with a convolutional neural network enables end-to-end training in the presence of weak image-level labels. We developed a new method for aggregating predictions from smaller regions of the image into an image-level classification by using the quantile function. The quantile function provides a more complete description of the heterogeneity within each image, improving image-level classification.
Results: We validated our method on five different classification tasks for breast tumor histology and demonstrated a visualization method for interpreting local image classifications. Further, we examined predicted biomarker heterogeneity across the entire test set to show how our method may lead to future insights into tumor heterogeneity.
Additional Applications: Our methods can be applied whenever there are weak annotations for a concept due to the time or expense required for creating finer-grained annotations or due to the inability of humans to perform such an annotation task. More broadly, intra-class heterogeneity is present in many other classification problems from environmental to economic. Any time a label is applied to a group of cells, people, households, organisms, or other potentially diverse components, some amount of intra-class heterogeneity is present. The methods that we developed could provide insight into the amount of heterogeneity in a data set and, from that, a better understanding of the data itself.

Breast Cancer Recurrence
Background: Pathologists examine tissue to identify the presence of a tumor and to assess tumor aggressiveness, the accuracy of which is fundamental in providing appropriate and timely treatment. Better survival prediction can help to determine how closely to monitor patients and which patients should be offered entrance into clinical trials. This project used histology image features and gene expression data to predict early recurrence of breast cancer.
Solution: Both linear and deep methods were explored as a means for prediction. The simplest models were binary and predicted recurrence at fewer than three years or not. More complex formulations used the Cox Proportional Hazard model in both a linear and a deep learning framework.
Alternative Applications: While this project studied cancer recurrence, the same models apply for survival. Further, any application to predict time to outcome - such as time to death or time to failure - can make use of these models.
Integration of Image and Genomic Features of Breast Cancer
Background: Histologic image features and genomics provide two complementary views of tumors. By combining the two, a more complete picture of tumor prognosis and treatment models is possible. Each alone has been shown to inform certain decisions made by doctors, but an integrated model can provide a more predictive analysis. We used these tumor features to predict genomic subtype and grade.
Challenge: The breast cancer data set was small with only around 1,000 samples. This is typical for medical data and brings challenges for many machine learning tasks. In particular, the deep learning models that excel with large training sets tend to overfit. Further, most commonly-used multimodal analysis techniques are unsupervised - they make no use of class labels. Our goal was to create a method to find a shared embedding of the input modalities that is also discriminative.
Solution: We created a new set of deep learning techniques based off a popular data analysis technique called Canonical Correlation Analysis (CCA). CCA projects a pair of modalities into a shared space. While CCA is often used for classification tasks, we found that recent deep learning variants of CCA found many features that were of no use for classification. Our methods resolve this by ensuring that the shared space is also discriminative.
Results: We tested our methods for classifying breast tumors when both modalities are available for training but only one is available at test time, as well as two other applications. Our results showed a large increase in classification accuracy over previous methods and increased robustness to small training set sizes. We also demonstrated utility for semi-supervised learning when labels were only available for some samples.
Additional Applications: The robustness properties of our methods make them especially suitable for medical applications in which patient samples are limited. Parallel modalities of data are increasingly common in many other applications: other imaging modalities, images and text, audio and video, parallel texts of different languages, or even different feature representations for the same data. Beyond classification tasks, CCA-based methods can also be used for clustering or any other task-driven goal that can be used in a deep network.
Hierarchical Task-Driven Feature Learning for Tumor Histology
Background: Most automated analysis of histology follows a general pipeline of first segmenting nuclei, then characterizing color, texture, shape, and spatial arrangement properties of cells and nuclei. These hand-crafted features are time-consuming to develop and do not adapt easily to new data sets. More recent work has begun to learn appropriate features directly from image patches. This project was completed before deep learning tool kits were readily available and instead used dictionary learning as a means for learning representations from image patches.
Solution: Through learning small and large-scale image features, we captured the local and architectural structure of tumor tissue from histology images. This was done by learning a hierarchy of dictionaries, where each level captured progressively larger scale and more abstract properties. By optimizing the dictionaries further using class labels, discriminating properties of classes that are not easily visually distinguishable to pathologists were captured.
Results: We explored this hierarchical and task-driven model in classifying malignant melanoma and the genomic subtype of breast tumors from histology images. We also showed how interpreting our model through visualizations can provide insights to pathologists.
Additional Applications: Dictionary learning can be used as a discriminative representation for classification of textures, objects, scenes, or many other types of images. It can also form a compact descriptor for image retrieval. While not as powerful a discriminator as many deep learning methods, it is much more interpretable and so would be a suitable representation when insight into the features driving classification is important.

Human Retina Segmentation from Optical Coherence Tomography
Background: The human retina is made up of ten layers. Measuring the thickness of these layers is an important step in detecting diseases such as macular degeneration, glaucoma, and diabetic retinopathy. Optical Coherence Tomography (OCT) is a non-invasive technique to generate cross-sectional imagery of ocular structures, enabling study of individual retina layers.
Solution: This project developed a solution for segmenting layers of the human retina from noisy OCT images. Retina layers were detected incrementally, with the boundary for each delineated while smoothly maneuvering around noisy pixels. The accuracy for each layer was measured during each step of the project and failure modes studied in order to discover areas for improvement.
Additional Applications: The active contour technique used for a smooth segmentation can be adapted to many other image segmentation applications. It especially excels in noisy images and can be tuned for different levels of smoothing.
Improved Precision of Mouse Retina Segmentation from Optical Coherence Tomography
Background: The goal of this project was to improve the precision of a 3D segmentation algorithm that was challenged by noise and outliers. 2D frames were being processed independently, contributing to some of the noise. Analysis revealed by 3D plots showed the presence of background noise from the imaging device, structured noise caused an oscillation due to breathing, and outlying points near the edge of the scan and around internal structures. A solution to handle these complications also needed to run in a reasonable time.
Solution: A sequence of steps was developed that included aligning adjacent slices, robustly removing outliers, interpolating missing points, smoothing the segmentation, and cropping the bounds of the retina. A functional prototype was produced, and the segmentation accuracy was measured before and after implementation to verify the improvements.
Additional Applications: The techniques used in this project can applied as post-processing whenever an initial algorithm produces an excessively noisy output. Understanding the modalities of noise enables the best selection of noise reduction and clean up techniques.
Keyword Prediction from Movie/TV Series Metadata
Background: With the increasing amount of video content available to consumers, methods to assist in finding relevant content are of increasing importance. By labeling with keywords describing different aspects of the content, such as the setting or theme, movies can be sorted or searched with these new criteria. Labeling the descriptive keywords by hand is, however, both time-consuming and subjective.
Solution: This project developed and analyzed a method for predicting keywords for a movie or TV series automatically using commonly available descriptive metadata such as the cast list, genres, a plot description, and reviews. It can be used to assign keywords for an unlabeled movie or to find additional keywords for an already labeled movie.
Additional Applications: Many techniques employed in this project are suitable for other tasks, including: handling structured and unstructured input data, incorporating multiple modalities of data, and imputation of missing data.
Mood: endearing, engaging, incredible
Setting: california, small town
Subject: daunting task, romance
Theme: adventure, discovery, quest
Time period: 1950s, 1980s
Mood: charming, heartwarming, witty
Setting: australia
Subject: abduction, daunting task, father/son relationship, friendship
Theme: escape, growing up, quest, underdog
Time period: 2000s
Texture Methods for Histology Image Analysis
Background: Recent advances in digitizing whole microscope slides have enabled the creation of large databases of images. Examining each image by hand to determine the composition of different tissue types and inspect for anomalies is both inefficient and subjective, even for a trained pathologist. With the development of image analysis techniques, the work can be automated and quantitative measures standardized. This project explored texture methods for classifying tissue types in histology images. Such images are taken by a microscope of tissue, such as that obtained by biopsy. Prior to imaging, sections of tissue are stained to increase the contrast of structures of interest. Hematoxylin and eosin (H&E) is one such stain which turns nuclei blue and cytoplasm pink. The magnification is high enough that cell structures such as nuclei and membranes are visible and different tissue types can be identified.
Solution: Through the use of color and texture analysis methods, the classification of tissue types was automated. The data set used in this work consisted of breast tissue that had been stained with H&E. Three different tissue types of interest were labeled and used to develop a classifier that could later be used for segmentation. This project explored color and texture methods for characterizing the appearance of tissue. The standard approach to texture classification involves convolving the image with a set of filters and clustering the responses into textons. This technique was compared with a variation involving quantile functions and methods to measure local image curvature.
Additional Applications: The texture methods used in this project can be used for classifying or segmenting any type of image. In a later project they were used to initialize a tissue segmentation routine.

Scene Classification of Images and Video via Semantic Segmentation
Background: Scene classification is used to categorize images into different classes, such as urban, mountain, beach, or indoor. This project tackled scene classification of television shows and feature films. These types of media bring unique challenges that are not present in photographs, as many shots are close-ups in which few characteristics of the scene are visible.
Solution: The video was first segmented into shots and scenes, and key frames from each shot were analyzed before aggregating the results. Each key frame was classified as indoor or outdoor. Outdoor frames were further broken down by a semantic segmentation that provided a label to each pixel. These labels were then used to classify the scene type by describing the arrangement of scene components with a spatial pyramid. The predicted keyframes labels were then summarized for each shot and scene.
Results: We tested our method on a large database of videos and compared with prior work on photographs. Validation of the semantic segmentation was shown on a set of hand-labeled images. Our work improved the semantic segmentation and scene classification of images and, to the best of our knowledge, was the first working system on video.
Additional Applications: The methods developed in this project are applicable when classification of an image depends on the proportion of its components. Semantic segmentation is now commonly used for self-driving cars and other autonomous navigation applications. Classifying the overall content of an image or video sequence can provide keywords for search and retrieval.

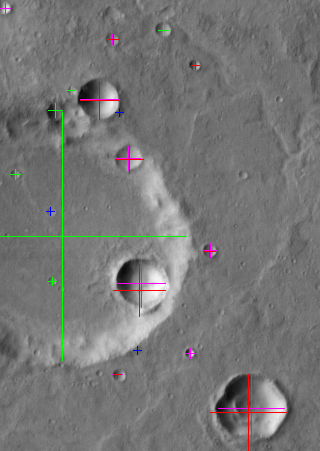
Crater Detection on Mars
Background: Detecting craters in satellite images has many applications for planetary geologists, including providing an indicator of the age of the terrain and in registering images taken in different modalities. Craters are so numerous that automated methods can greatly increase the efficiency of this analysis.
Solution: The method developed in this project started with identifying candidate craters by locating circular features. A set of intensity, texture, shading, template, and boundary features were then extracted on each candidate to learn a model of a crater. This model was successfully used to distinguish craters from non-craters.
Alternative Applications: Many modern object detection algorithms start by proposing candidate regions, followed by a second stage to determine whether each region contains the desired object. By finding the candidate regions efficiently and with high recall, the second stage can provide the necessary precision to the algorithm. This combination makes for an efficient and accurate algorithm that is applicable for many types of objects.
Rock Detection and Mapping from Satellite Imagery
Background: This project used high resolution images taken by the HiRISE camera on board the Mars Reconnaissance Orbiter. HiRISE produces images that are 20,000 pixels across with a resolution as high as 30 cm/pixel, enabling us to see boulder-sized rocks on the surface. Such images were used to decide on a safe landing location for another Mars explorer, the Phoenix lander.
Solution: The rocks in question appear as a light spot (the rock) adjacent to a dark region (a shadow). Dependent upon the size of the shadow and given that the sun position is known, the approximate size of the rock can be calculated. Potential landing locations for Phoenix were represented by large ellipses with an area of hundreds of square kilometers. Rock distribution maps were computed over these areas to show the density of rocks, providing the Phoenix team with information about potential hazards in each region.
Alternative Applications: The rock detection technique used on this project was quite simple but enabled scaling to very large images. Many microscopic or high resolution applications deal with similarly large images and only require simple processing of image components in order to produce a large-scale summary map.

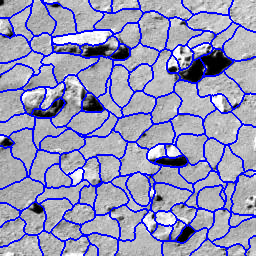
Automatic Rock Detection and Classification in Natural Scenes
Background: Autonomous geologic analysis of natural terrain is an important technique for science rovers exploring remote environments such as Mars. By automatically detecting and classifying rocks, rovers can efficiently survey an area, summarize results and concentrate on unusual discoveries, thus maximizing the scientific return. This capability is fundamental in enabling robots to conduct scientific investigations autonomously.
Solution: In this project, techniques for characterizing rocks using albedo, color, texture, and shape properties were developed. These and other features were used to detect rocks, using a superpixel segmentation algorithm to locate and delineate the rocks with an accurate boundary. Once segmented, a Bayesian procedure was applied to geologically classify rocks.
Results: Experiments demonstrated the usefulness of the albedo, color, texture, and shape measures. The high performance of the rock detection and segmentation algorithm was analyzed and the results of geologic rock classification assessed. The methods successfully performed geologic rock analysis, while giving an indication of what features were most important for this and similar tasks.
Alternative Applications: As color, texture, and shape are such powerful features, this approach is also applicable to detecting, segmenting, and classifying other natural objects and terrain features. Similar techniques can be used in natural settings on land and under water where characterization of context is important to robot behavior.

Multi-scale Features for Detection and Segmentation of Rocks in Mars Images
Background: Geologists and planetary scientists can benefit from methods for accurate segmentation of rocks in natural scenes. However, rocks are poorly suited for current visual segmentation techniques - they exhibit diverse morphologies and have no uniform property to distinguish them from background soil.
Solution: We addressed this challenge with a novel detection and segmentation method incorporating features from multiple scales. These features included local attributes such as texture, object attributes such as shading and two-dimensional shape, and scene attributes such as the direction of illumination. Our method used a superpixel segmentation followed by region-merging to search for the most probable groups of superpixels. A learned model of rock appearances identified whole rocks by scoring candidate superpixel groupings. We evaluated our method’s performance on representative images from the Mars Exploration Rover catalog.
Alternative Applications: Many natural objects and biological structures exhibit similarly diverse morphologies and may also benefit from an integrated detection and segmentation routine.
Rock Shape Characterization
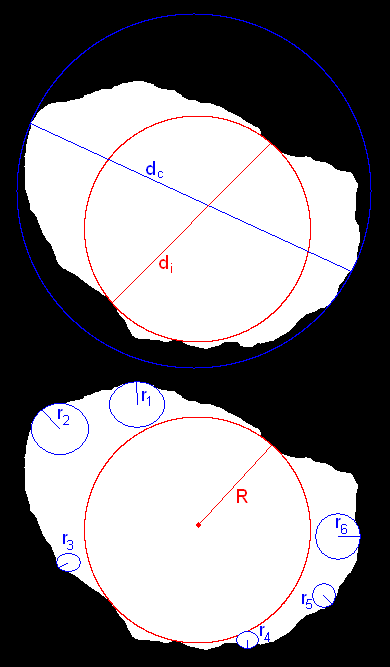
Background: When examining rocks and sediments found in the field, geologists find shape to be an important property to characterize. Shape properties can be divided into three categories: form, roundness and texture. Form characterizes the overall shape of a rock, such as how close to spherical it is. Roundness measures the extent to which the edges have been smoothed out. Texture describes the markings on the surface of the rock, such as striations. The shape of a rock can help determine the history of it, such as where it came from and how it was transported. For example, rocks transported in water tend to get worn down by the force of the water, resulting in a more rounded surface, while the overall shape of the rock changes little after it breaks off from an outcrop. Computing metrics to describe these characteristics can aid in the autonomous classification of rocks. For example, by computing the shape, albedo and texture of a rock, it could be classified as sandstone, limestone, marble, or slate.
Solution: This project explored possible shape descriptors beyond the standard sphericity and roundness that characterize properties of rocks and might be useful features in geologic classification. Each measure was computed on a small set of rocks and the results were analyzed.
Alliterative Applications: Shape properties can be important for classifying natural objects and detecting manufacturing anomalies. While sphericity and roundness are specific to the geologic community, other custom shape characterizations can be developed for unique applications.
 Source:
Source: