A review of techniques for modeling whole slide histology images and recommendations for different situations.
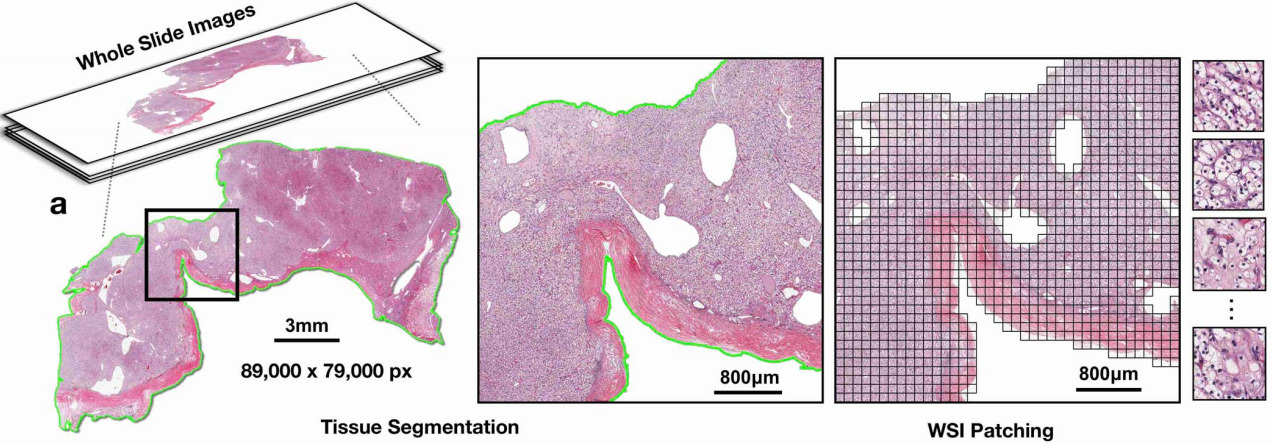
Whole slide histopathology images can be as large as 100,000 pixels across. Such massive images are both time-consuming and costly to annotate in detail. For some tasks, pathologists can annotate individual features within the image like tissue types or individual structures like mitotic figures. But for other tasks, only higher-level annotations are possible.
Patient-level labels could be obtained from clinical data, such as whether a pre-invasive lesion became invasive, how long the patient lived after diagnosis, or whether they responded to a particular treatment. It could also be a label obtained from a different type of analysis performed on the tumor, such as molecular analyses to identify mutations or genomic subtype. Alternative methods of immunohistochemical staining could also produce a label for the entire tumor, for example the receptor status.
Recent research has demonstrated that deep learning algorithms have the ability to predict these types of patient-level attributes from H&E whole slide images (WSIs). Because the images are so large and no a-priori knowledge of which patches within them are associated with the label, this is known as weakly supervised learning. It is the job of the algorithm to figure out which patches within the WSIs are relevant to the prediction task.
Gigapixel images are too large to fit on a GPU all at once; they are typically broken into smaller patches for training the deep learning model. This article will look at how discriminative features can be learned from these smaller patches and how they can be used to predict slide- or patient-level attributes. More recent innovations have also found ways to train a CNN on the WSI – but with some remaining limitations that will be discussed towards the end of this article.
There are two common tricks that enable WSIs to be processed on a GPU: sampling a subset of image patches or downsizing the image representation.
Patch Sampling
The entire WSI cannot fit in GPU memory at once, so one solution is to select a subset of patches from the image. The following patch selection approaches have been employed:
Random selection: The simplest way to sample patches from WSIs images is randomly. During each epoch of training, a different random subset of patches is selected from each WSI [Naik2020]. If the quantity and size of patches is sufficiently small, then it will fit on the GPU.
Identify tumor, then random selection: Either a pathologist can annotate tumor regions [Zhu2016] or a cancer detection algorithm can be trained on annotated tumor and non-tumor patches to identify tumor in new WSIs. Then a subset of tumor patches can be randomly selected each epoch.
Cluster image patches: To capture a more diverse view of tissue appearance, image patches can be clustered. This may be done by clustering the patches in each image individually into, say, 10 clusters. Then one patch from each cluster will be sampled, and these 10 patches together will represent the WSI [ Yao2020]. Another variation on this is to cluster patches across the entire training set and assign the closest patch to the cluster centroid in each WSI to that cluster. Xie et al. applied this clustering method iteratively, recomputing the cluster centroid after each epoch of training [Xie2020].
Patch Compression
The alternative to subsampling image patches is to use the entire WSI but in a compressed representation. Once again, multiple strategies have been applied for encoding image patches into a smaller representation.
A CNN can be trained on another dataset and used to encode image patches. Often CNNs pre-trained on ImageNet are transferred to histology images [Lu2021]; however, the same technique can be more effective when pre-trained on images from another histology dataset [Tellez2020].
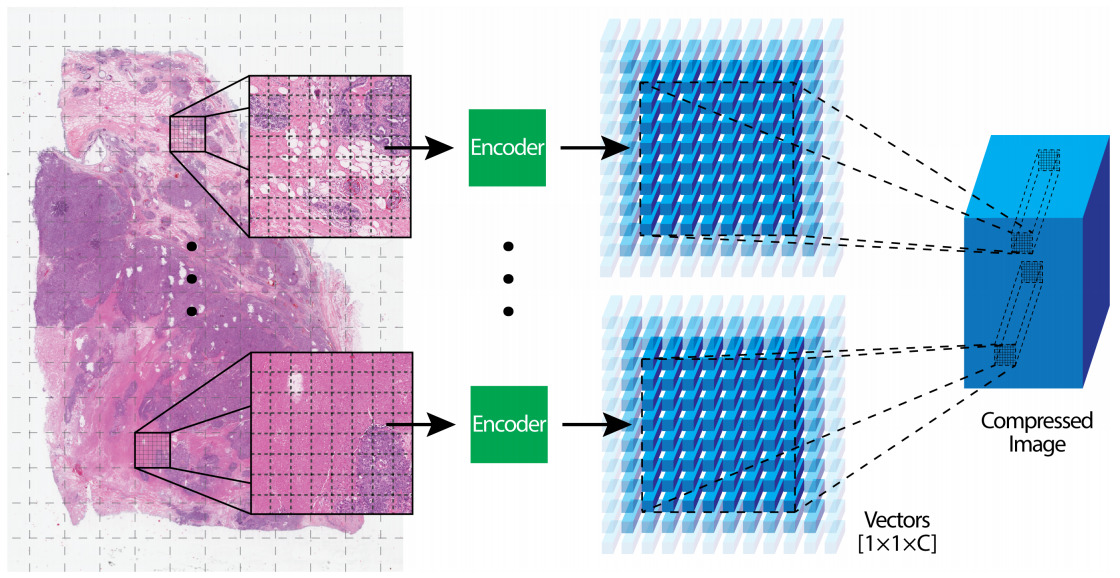
Unsupervised, self-supervised, and Generative Adversarial Network (GAN) approaches to pre-training have also shown success [Tellez2019, Dehaene2020].
Aggregating Patches
Feature encodings from multiple patches in a WSI must then be aggregated to predict the class. These aggregation methods may be used in training an end-to-end CNN with a subset of image patches or as a second stage model to operate on patches that have already been compressed to a smaller representation.
Most commonly this is done with the multiple instance (MI) learning paradigm. Each WSI is a bag that has multiple instances (image patches). We know the label for the bag but not the labels for the instances.
Aggregation may be performed on patch features or patch predictions. For aggregating patch predictions, traditional MI learning defined a positive bag as having one or more positive instances and a negative bag as one with all negative instances. This definition works great for tasks such as cancer detection, where even a small amount of malignant tissue is considered cancer. If the training dataset is sufficiently large, the model can learn from this simple MI definition. Campanella et al. used 44k biopsy WSIs to train their model [Campanella2019].
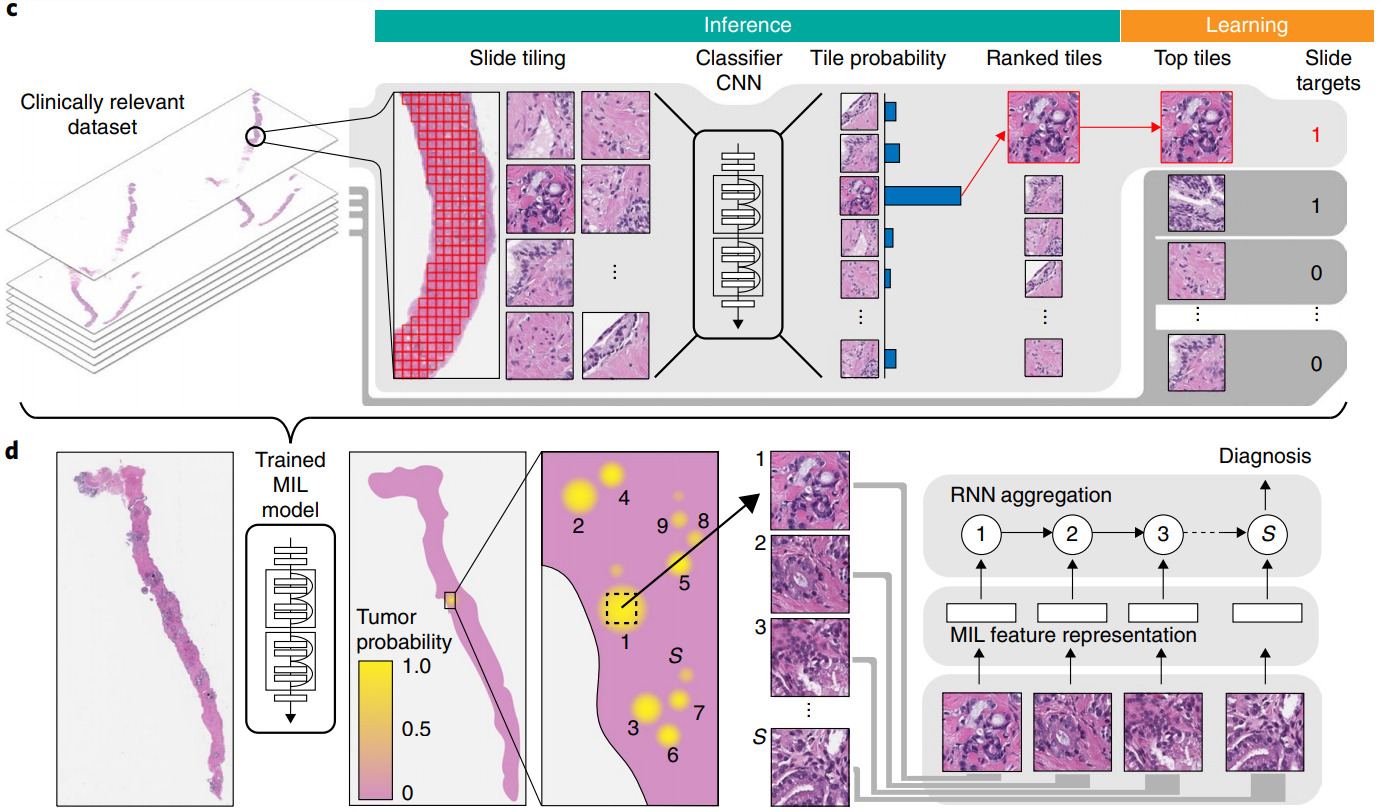
For other tasks, a different method of aggregating patch predictions may be more appropriate – for example, a majority vote of class predictions or averaging the prediction scores across the patches. Some of my prior work computed a quantile function of the patch prediction scores to more fully characterize their distribution, then trained a linear classifier from the quantile function [Couture2018a] or built quantile aggregation into a CNN [Couture2018b].
Aggregating Patch Features
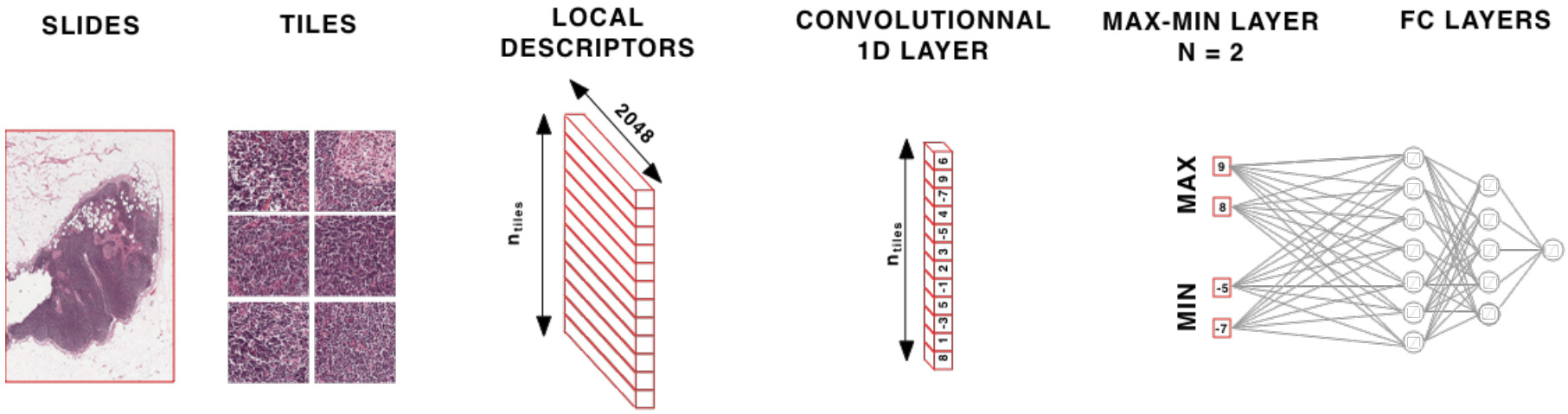
Aggregating patch features is an alternative approach. Once aggregated, fully connected layers may be applied on top before the final prediction. There are a variety of options for feature aggregation: max or mean pooling (taking the max or mean value across the patch encodings for each feature) [Wulczyn2020], identifying the most informative patches, or a weighted sum.
Deep learning can be used to identify the most informative patches in a WSI. Courtiol et al. created a max-min layer to identify top patches and negative evidence [Courtiol2018]. They found that while mean pooling works well for diffuse disease, the max-min layer provides a solid boost when the cancer is not diffuse.
A more general formulation is the self-attention mechanism that can learn patch weights that are used to compute a weighted sum of patch embeddings [ Yao2020, Lu2021, Mobadersany2021]. This is now the most prevalent method for aggregating patch features.
Self-attention uses the patch encoding to calculate a weight for each patch. The patch features are then aggregated as a weighted sum. This formulation enables the model to learn which patches are most informative for a particular task and weight the features from those patches more highly.
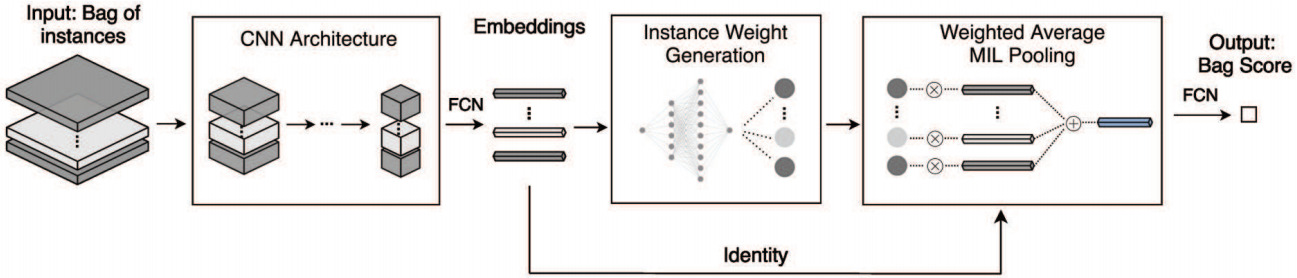
Taking self-attention models a step further, Li et al. used the computed patch weights to identify hard negative patches [Li2019]. From that, they constructed hard negative bags that are used to augment training.
Another variant of self-attention is certainty pooling, where certainty is calculated as the inverse standard deviation of instance Monte Carlo dropout predictions [Gildenblat2020]. This measure of model certainty was shown to be more explainable and robust than alternatives like self-attention when only a small number of patches contain evidence for the desired task.
Both the self-attention and certainty pooling weighting schemes provide some interpretability. The weights can be overlaid on the WSI as a heatmap showing the parts of the image that are most informative for the prediction task.
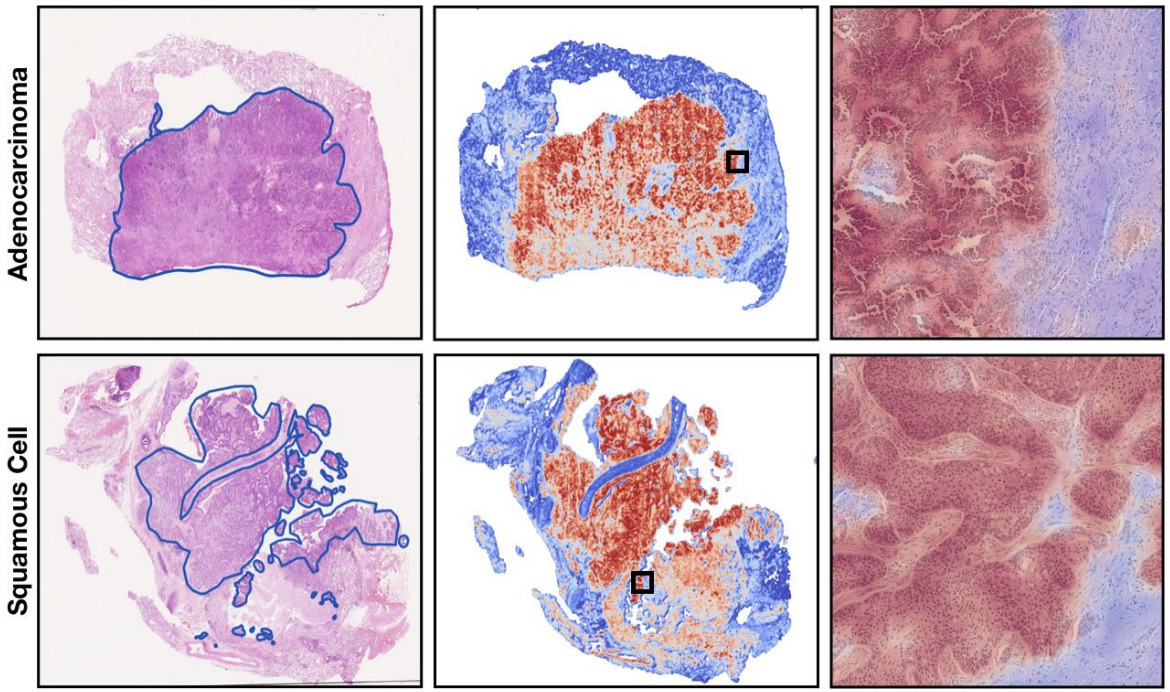
Tricks to Achieve End-to-End Training
While a WSI cannot fit on a single GPU, some tricks have enabled a WSI to be processed as if it were all held in GPU memory.
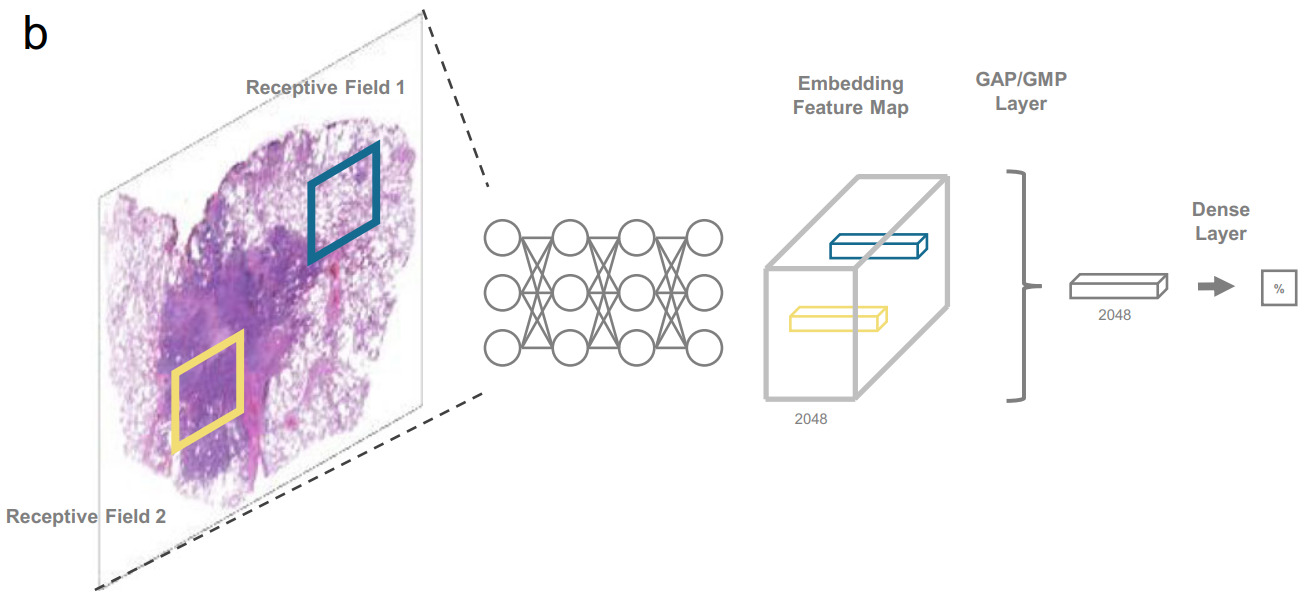
A CUDA feature called unified memory provides the GPU direct access to host memory. Similar to virtual memory, pages are swapped on the GPU when requested. Through this technique, Chen et al. were able to process images as large as 20k x 20k pixels [Chen2021]. Any larger became prohibitively slow. To accommodate larger WSI, they downsized them by a factor of 4 in each dimension. This technique is best suited for easier tasks that can operate on lower magnification images.
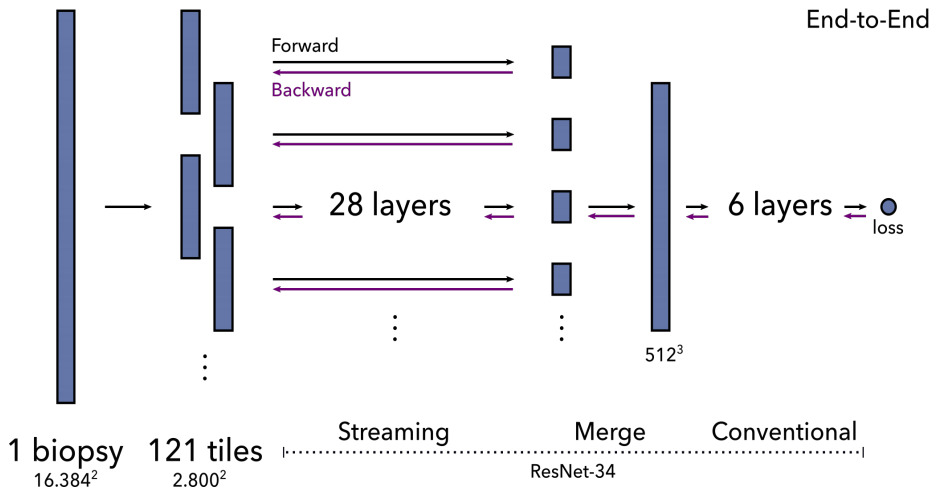
An alternative is a method called streaming that exploits the locality of most CNN operations. It combines precise tiling and gradient checkpointing to reduce memory requirements. To stream the forward pass of a CNN, you first calculate the feature map of a chosen layer in the middle of the network. This layer is smaller than the original image because of downsampling, so fits on the GPU. This reconstruction of the intermediate feature map is then fed to the remainder of the network. The backward pass is computed in a similar fashion.
Pinckaers et al. first demonstrated this technique with a small and simple CNN on 8k x 8k images [Pinckaers2020a]. They subsequently showed it for a ResNet on 16k x 16k images [Pinckaers2020b]. The limitation of streaming is that it cannot handle feature map-wide operations such as batch normalization in the streaming (lower) part of the network. As a workaround, they froze the mean and variance of batch normalization layers.
Yet another possibility is a spatial partitioning approach called halo exchange [Hou2019]. Hou et al. demonstrated this for segmentation of 512x512x512 CT images and speculated that it would also be a good fit for histopathology. This technique distributes the input and output of convolutional layers across GPUs with the devices exchanging data before convolutional operations.
From unified memory to streaming to halo exchange, each of these approaches enables end-to-end training of much larger images – but still with current limits around 20k x 20k pixels or less. We are not yet able to process larger WSIs without downsizing the images and losing details that may be important for prediction.
Alternative Techniques for Weak Supervision
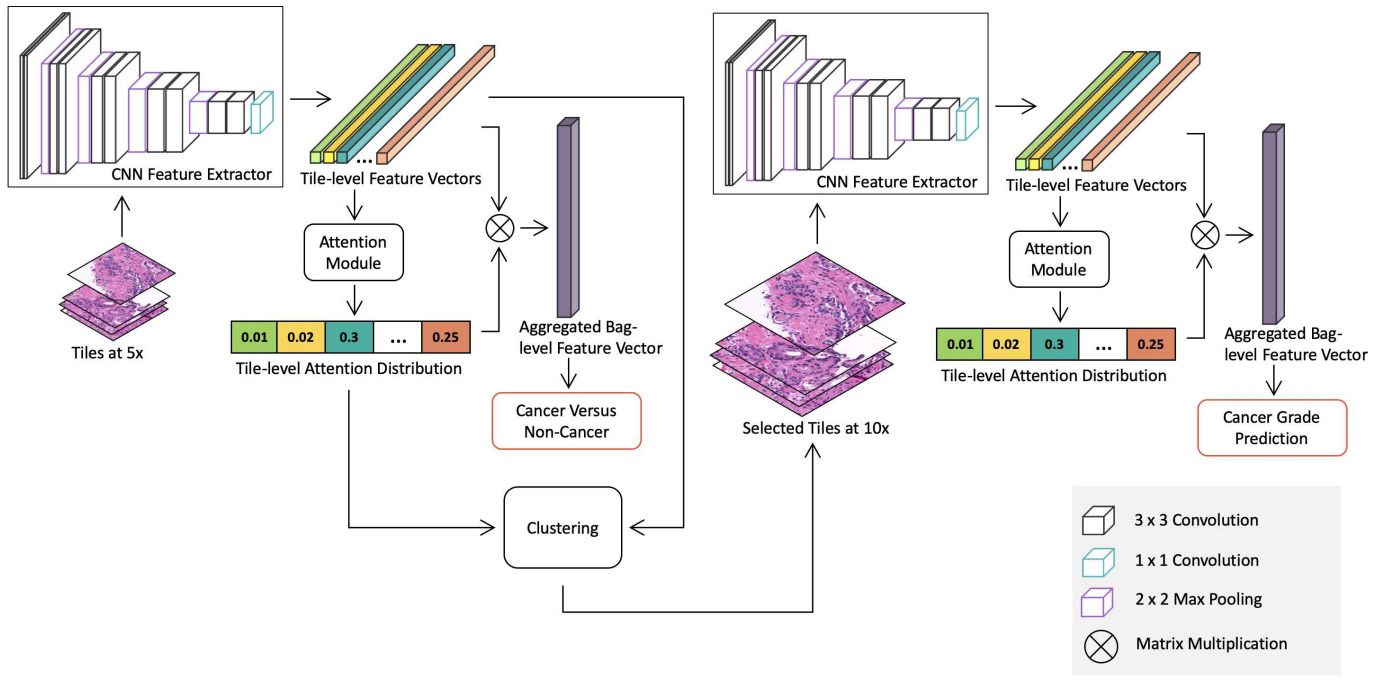
Some methods use a combination of the above techniques. Li et al. downsized WSIs to 5x magnification, used clustering to capture variations in patch appearance, and an attention model to identify important clusters (and the patches within them) [Li2021]. After using this method to detect cancer, they applied the same approach at 10x magnification with only the patches identified as cancer to predict grade.
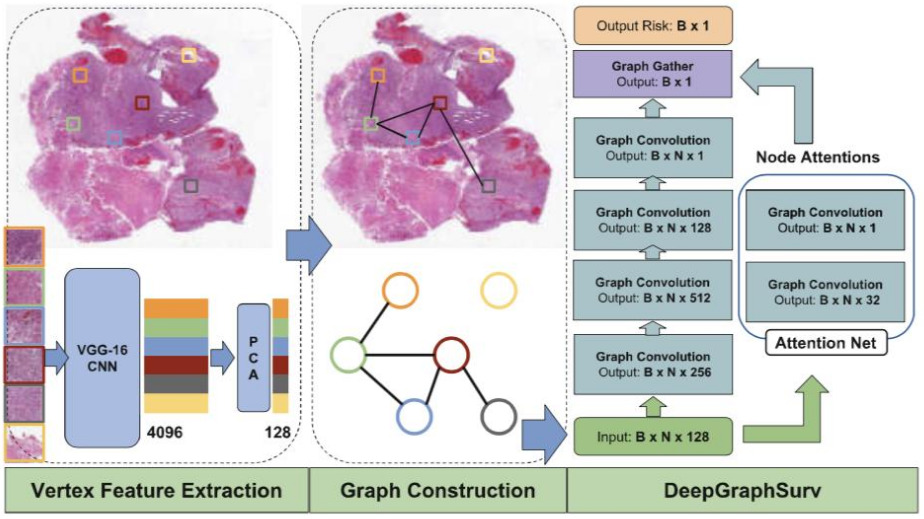
The above applications to WSIs all use a CNN to model the images. A graph neural network provides an alternative representation. A CNN is applied to image patches, then they are connected into a graph neural network that learns a representation for the WSI in order to make predictions [Li2018, Levy2021]. The graph captures relationships between neighborhoods of patches and other potentially important architectural features.
Wrap Up and Recommendations
So which weak supervision approach should you choose? It likely depends on your dataset. For example, if you have tens of thousands of images, you might be able to get away with a traditional MI approach with a CNN to detect cancer [Campanella2019]. But most WSI datasets are not that large.
Can you exclude parts of the image, such as non-tumor regions? That reduces the amount of image data to be processed – but could exclude regions that are potentially informative, like for a survival model.
Another perspective is to identify whether tissue architecture or small scale image features are more likely to be most informative for your task. If the former, the unified memory or streaming methods on downsized WSI would be good places to start. Or graph neural networks. But if you need higher magnifications to learn appropriate features, try out either the patch sampling or patch compression approaches combined with an aggregation method like self-attention. These methods can even work well with fewer than 1000 images for training.
If you think a good CNN representation can be transferred from another dataset or learned in a self-supervised way, then try a patch compression approach. For the most challenging tasks, like predicting survival or other outcomes, you might need to provide direct supervision on the image patches. In this case, patch sampling combined with attention might be your best bet.
These are merely speculations though. Experiment with a couple of approaches that seem most appropriate for your dataset and task. Learn and repeat. This is science after all.
Want to receive regular computer vision insights for pathology delivered straight to your inbox?
Sign up for Computer Vision Insights
References
[Campanella2019] G. Campanella, M.G. Hanna, L. Geneslaw, A. Miraflor, V.W.K. Silva, K.J. Busam, E. Brogi, V.E. Reuter, D.S. Kilmstra, T.J. Fuchs, Clinical-grade computational pathology using weakly supervised deep learning on whole slide images (2019), Nature Medicine
[Chen2021] C.-L. Chen, C.-C. Chen, W.-H. Yu, S.-H. Chen, Y.-C. Chang, T.-H., M.Hsiao, C.-Yu. Yeh, C.-Y. Chen (2021), An annotation-free whole-slide training approach to pathological classification of lung cancer types using deep learning, Nature Communications
[Courtiol2018] P. Courtiol, E.W. Tramel, M. Sanselme, G. Wainrib, Classification and disease localization in histopathology using only global labels: A weakly-supervised approach (2018), arXiv preprint arXiv:1802.02212
[Couture2018a] H.D. Couture, L.A. Williams, J. Geradts, S.J. Nyante, E.N. Butler, J.S. Marron, C.M. Perou, M.A. Troester, M. Niethammer (2018), Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype (2018), npj Breast Cancer
[Couture2018b] H.D. Couture, J.S. Marron, C.M. Perou, M.A. Troester, M.Niethammer, Multiple Instance Learning for Heterogeneous Images: Training a CNN for Histopathology (2018), Medical Image Computing and Computer Assisted Interventions
[Dehaene2020] O. Dehaene, A. Camara, O. Moindrot, A. de Lavergne, P. Courtiol, Self-Supervision Closes the Gap Between Weak and Strong Supervision in Histology (2020), arXiv preprint arXiv:2012.03583
[Gildenblat2020] J. Gildenblat, I. Ben-Shaul, Z. Lapp, E. Klaiman, Certainty Pooling for Multiple Instance Learning (2020), arXiv preprint arXiv:2008.10548
[Hou2019] L. Hou, Y. Cheng, N. Shazeer, N. Parmar, Y. Li, P. Korfiatis, X. Song, High resolution medical image analysis with spatial partitioning (2019), arXiv preprint arXiv:1909.03108
[Levy2021] J. Levy, C. Haudenschild, C. Bar, B. Christensen, L. Vaickus, Topological feature extraction and visualization of whole slide images using graph neural networks (2021), BioComputing
[Li2018] R. Li, J. Yao., X. Zhu, Y. Li, J. Huang, Graph CNN for survival analysis on whole slide pathological images (2018), Medical Image Computing and Computer Assisted Intervention
[Li2019] M. Li, L. Wu, A. Wiliem, K. Zhao, T. Zhang, B. Lovell, Deep instance-level hard negative mining model for histopathology images (2019), Medical Image Computing and Computer-Assisted Intervention
[Li2021] J. Li, W. Li, A. Sisk, H. Ye, W.D. Wallace, W. Speier, C.W. Arnold, A multi-resolution model for histopathology image classification and localization with multiple instance learning (2021), Computers in Biology and Medicine
[Lu2021] M.Y. Lu, D.F. Williamson, T.Y. Chen, R.J. Chen, M. Barbieri, F. Mahmood, Data-efficient and weakly supervised computational pathology on whole-slide images (2021), Nature Biomedical Engineering
[Mobadersany2021] P. Mobadersany, L.A.D.Cooper, J.A. Goldstein, GestAltNet: aggregation and attention to improve deep learning of gestational age from placental whole-slide images (2021), Laboratory Investigation
[Naik2020] N. Naik, A. Madani, A. Esteva, N.S. Keskar, M.F. Press, D. Ruderman, R. Socher (2020). Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains (2020), Nature Communications
[Pinckaers2020a] J.H.F.M. Pinckaers, B. van Ginneken, G. Litjens, Streaming convolutional neural networks for end-to-end learning with multi-megapixel images (2020), IEEE Transactions on Pattern Analysis and Machine Intelligence
[Pinckaers2020b] H. Pinckaers, W. Bulten, J. van der Laak, G. Litjens, Detection of prostate cancer in whole-slide images through end-to-end training with image-level labels (2020), arXiv preprint arXiv:2006.03394
[Tellez2019] D. Tellez, G. Litjens, J. van der Laak, F. Ciompi, Neural image compression for gigapixel histopathology image analysis (2019), IEEE Transactions on Pattern Analysis and Machine Intelligence
[Tellez2020] D. Tellez, D. Höppener, C. Verhoef, D. Grünhagen, P. Nierop, M. Drozdzal, F. Ciompi, Extending Unsupervised Neural Image Compression With Supervised Multitask Learning (2020), Medical Imaging with Deep Learning
[Wulczyn2020] E. Wulczyn, D.F. Steiner, Z. Xu, A. Sadhwani, H. Wang, I. Flament-Auvigne, C.H. Mermel, P.-H.C. Chen, Y. Liu, M.C. Stumpe, Deep learning-based survival prediction for multiple cancer types using histopathology images (2020), PLoS ONE
[Xie2020] C. Xie, H. Muhammad, C.M. Vanderbilt, R. Caso, D.V.K. Yarlagadda, G. Campanella, T.J. Fuchs, Beyond Classification: Whole Slide Tissue Histopathology Analysis By End-To-End Part Learning (2020), Medical Imaging with Deep Learning
[Yao2020] J. Yao, X. Zhu, J. Jonnagaddala, N. Hawkins, J. Huang, Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks (2020), Medical Image Analysis
[Zhu2016] X. Zhu, J. Yao, J. Huang, Deep convolutional neural network for survival analysis with pathological images (2016), International Conference on Bioinformatics and Biomedicine