|
Hi ,
My webinar "Demystifying Foundation Models for Pathology" will be live tomorrow. This is your last chance to join me for game-changing insights in this rapidly-evolving domain.
📅 Tomorrow, January 29th
🕛 11 am EST
Don't miss this chance to:
-
Explore cutting-edge AI models transforming pathology
-
Learn how to analyze gigapixel whole slide images at scale
-
Discover real-world applications in cancer detection and diagnosis
Who should join?
-
Startup founders in healthtech
-
AI researchers pushing boundaries
-
Healthcare innovators seeking the next big breakthrough
Bonus: Get a 25% discount for an in-depth team workshop to help you get started using foundation models!
There's still time to be part of this game-changing discussion. Register now!
And if this topic doesn't catch your attention, don't worry, there will be a fresh one next month.
Heather
|
|
|
|
|
|
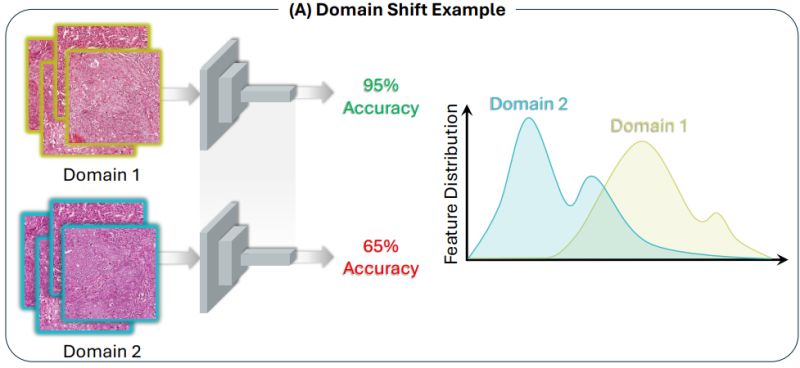 |
Research: Domain Generalization
Benchmarking Domain Generalization Algorithms in Computational Pathology
Distribution shifts like scanner differences and staining variations are a common cause of model performance degradation on inference images.
While a variety of domain generalization approaches have been developed, their evaluation on histology datasets has been pretty ad hoc.
Neda Zamanitajeddina et al. provide a comprehensive benchmark of 30 different algorithms on 3 different computational pathology tasks.
The evaluated state-of-the-art algorithms from the DomainBed collection, self-supervised learning (SSL) and pathology-specific techniques (stain augmentation and stain normalization).
SSL and stain augmentation consistently outperformed all other methods.
The stain normalization algorithm outperformed some of the DomainBed algorithms but was not as helpful as stain augmentation.
However, there was no single best algorithm for all experiments.
The authors concluded with the following recommendations:
1) Ensure that experiments are designed properly. E.g., no data leakage and use domain-level stratification of cases between the train and test sets.
2) Fine-tune a pretrained model (such as SSL) instead of learning from scratch or starting with ImageNet weights.
3) Use data augmentation, especially modality-specific techniques such as stain augmentation.
4) A combination of other algorithms can be investigated if performance is still not sufficient.
Code |
|
|
|
|
|
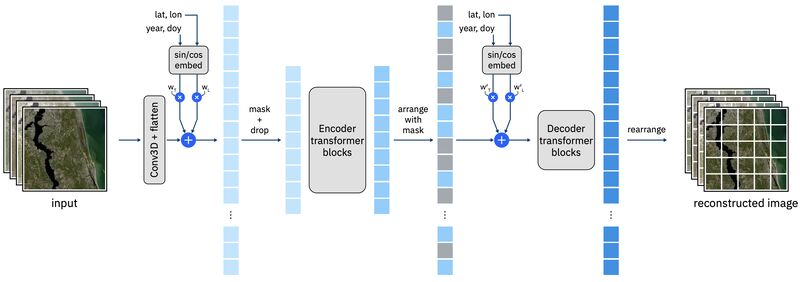 |
Research: EO Foundation Models
Prithvi-EO-2.0: A Versatile Multi-Temporal Foundation Model for Earth Observation Applications
Larger datasets and larger models are the standard approach for advancing vision foundation models.
As part of a collaboration between IBM, NASA, and Clark University, Daniela Szwarcman et al. released version 2 of Prithvi, a foundation model for Earth observation.
Prithvi-EO-2.0 was trained on 6-band Harmonized Landsat and Sentinel-2 imagery. They trained both 300M and 600M parameter vision transformer models, and a second set that also include temporal information.
While the original Prithvi was trained only on the US, this new version uses a globally diverse set of imagery.
They validated Prithvi-EO-2.0 on GEO-Bench, an extensive benchmarking dataset. It out-performed six other geospatial foundation models.
Further tests evaluated real-world applications in disaster response, land use and crop mapping, and ecosystem dynamics monitoring.
Models
Code |
|
|
|
|
|
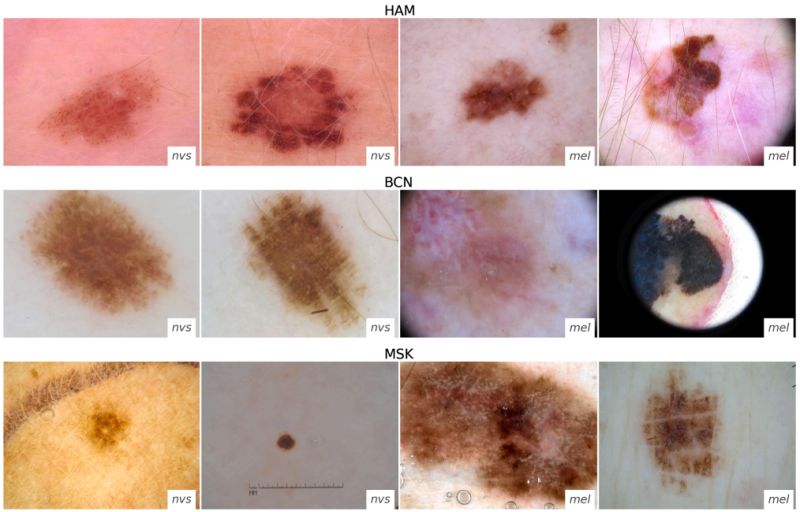 |
Research: Distribution Shifts
Domain shifts in dermoscopic skin cancer datasets: Evaluation of essential limitations for clinical translation
Domain shifts caused by technical or biological variations between the training and test set plague deep learning models for medical images.
Katharina Fogelberg et al. explored domain shifts for dermoscopic skin cancer.
They first collated three different datasets and broke them down by biological and technical shifts.
The technical shifts are due to different acquisition systems, while the biological shifts are most likely related to patient age and location on the body.
Using one dataset for training, they quantified and visualized the shift with each of the dataset groups.
To accommodate the domain shift, they experimented with a Domain Adversarial Neural Network (DANN), a form of unsupervised domain adaptation where the network has an additional classifier to predict the domain of an image. The network is tasked with its original goal, plus a gradient reversal module for the domain classifier to encourage the model to learn domain-invariant features.
DANN improved model generalization in 8 out of 10 of their test cases.
|
|
|
|
|
|
 |
Research: Distribution Shifts
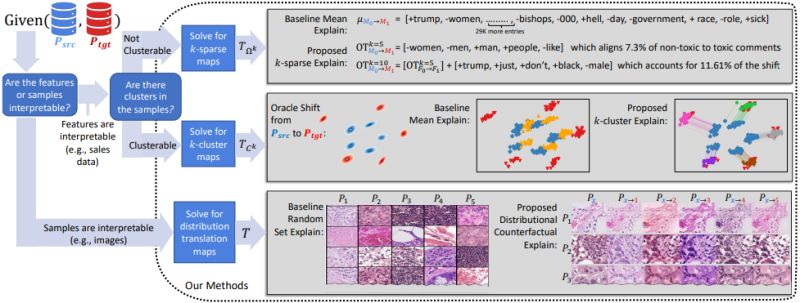
Towards Explaining Distribution Shifts
A distribution shift between the training set and inference can easily result in incorrect predictions from a model.
Understanding the distribution shift is key to evaluating what to do about it.
Sean Kulinski and David Inouye detailed three different approaches to explaining a distribution shift.
One of these approaches works well when the samples are interpretable (like images) and was demonstrated on a histopathology dataset.
The other two work for interpretable features and were demonstrated on two tabular datasets.
There are a number of other examples in the appendix.
|
|
|
|
|
|
|