 |
Research: Multimodal Pathology
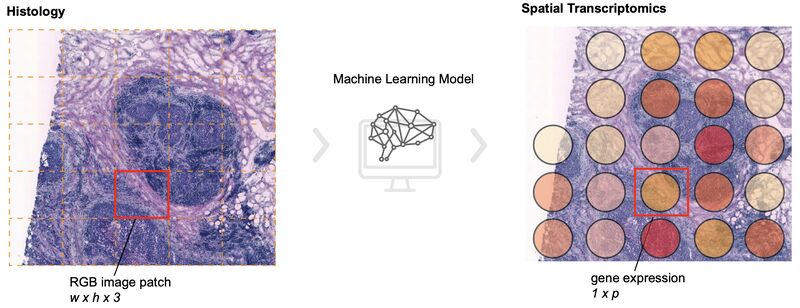
Benchmarking the translational potential of spatial gene expression prediction from histology
I've written a lot about approaches for predicting molecular properties of cancer over the past few years.
Most of these methods are focused on slide- or patient-level predictions of things like mutation status or gene expression.
But as the availability of more granular spatial information grows, so do the modeling opportunities.
Chuhan Wang et al. benchmarked a number of recent studies on predicting spatial gene expression from H&E.
They included assessments for different spatial resolutions and tissue types, plus they evaluated model robustness to image quality, generalizability, and the ability to predict patient survival using the inferred gene expression.
"Our overall observation is that the most complex deep-learning architectures involving more components were not superior in either SGE prediction or in translational potential categories. . . . [W]e found that methods effectively capturing both local and global features within single-spot patches, as exemplified by DeepPT, outperform methods attempting to incorporate additional spatial information from neighbouring spots. This observation suggests that SGE patterns in adjacent spots may not necessarily be strongly or consistently correlated. . . . Furthermore, including global information from the entire image slide may introduce noise since there is a large number of genes and some genes are not expressed across all spots. . . . Interestingly, transformer-based methods demonstrated better risk group stratification within HER2+ cancer subtype, suggesting that their ability to capture various histology characteristics could be more closely related to downstream clinical outcomes than SGE patterns."
These conclusions provide some insights into which approach to select, plus ideas for future research to include slide quality control and staining variations.
|
|
|
|
|
|
 |
Podcast: Impact AI
Foundation Model Series: Advancing Precision Medicine in Radiology with Paul Hérent from Raidium
Radiologists face a growing demand for imaging analysis, yet existing AI tools remain fragmented, each solving only a small part of the workflow. Today, we continue our series on domain-specific foundation models with Paul Herent, M.D., Msc, Co-Founder and CEO of Raidium. He joins us to discuss how foundation models could revolutionize radiology by providing a single AI-powered solution for multiple imaging modalities.
Paul shares his journey from radiologist to AI entrepreneur, explaining how his background in cognitive science and medical imaging led him to co-found Raidium. He breaks down the challenges of building a foundation model for radiology, from handling massive datasets to addressing bias and regulatory hurdles, and their approach at Raidium. We also explore Raidium’s vision for the future: its plans to refine multimodal AI, expand its applications beyond radiology, and commercialize its technology to improve patient care worldwide. Tune in to learn how foundation models could shape the future of radiology, enhance patient care, and expand global access to medical imaging!
|
|
|
|
|
|
Research: Multimodal Remote Sensing
On the Relevance of SAR and Optical Modalities in Deep Learning-based Data Fusion
Including multiple modalities of data in a model has the potential to improve over a single modality alone.
To do this successfully, you need to consider both your algorithmic approach for combining the modalities and your training data.
This article by Jakob Gawlikowski and Nina Maria Gottschling is a lesson in the latter -- your training data matters.
The authors investigated how the absence of cloudy samples in the training data affects the fusion of optical and SAR modalities in predicting land cover.
Previous models combining optical and SAR were trained on cloud-free datasets. However, the authors of this paper found that their performance dropped significantly on cloudy images -- even though SAR can see through clouds. Explainability techniques also showed that the models relied mostly on optical and did not use the benefits of SAR.
They trained alternative models that included cloudy pairs of optical and SAR images in the dataset.
These models performed significantly better on cloudy images and even showed some improvement for clear images.
Explainability techniques showed that the new models used the SAR modality more than the original models, especially on cloudy images.
This is partially a lesson in handling distribution shifts: your training set needs to include images from similar conditions to your inference data (e.g., clouds). But it is also a reminder to consider data-centric approaches to solving multimodal challenges.
|
|
|
|
|
|
 |
Insights: Foundation Models
Navigating Limited Data in Pathology AI: From Foundation Models to Fine-Tuning
A question from my recent webinar on foundation models for pathology: When training data is limited, how can we be sure that foundation model embeddings are generalizable?
Foundation models offer a promising solution for scenarios with limited labeled data in computational pathology. However, ensuring generalizability remains a critical challenge.
Here's a deep dive into strategies for maximizing performance with scarce labeled data:
1. 𝐅𝐨𝐮𝐧𝐝𝐚𝐭𝐢𝐨𝐧 𝐌𝐨𝐝𝐞𝐥 𝐀𝐝𝐯𝐚𝐧𝐭𝐚𝐠𝐞𝐬
Pre-trained on vast unlabeled datasets, these models reduce reliance on labeled data. They capture a broad spectrum of patterns in tissue morphology and staining characteristics.
2. 𝐅𝐢𝐧𝐞-𝐓𝐮𝐧𝐢𝐧𝐠 𝐒𝐭𝐫𝐚𝐭𝐞𝐠𝐢𝐞𝐬
Linear probing (tuning only the top layer) reduces overfitting risk.
3. 𝐕𝐚𝐥𝐢𝐝𝐚𝐭𝐢𝐨𝐧 𝐢𝐬 𝐂𝐫𝐮𝐜𝐢𝐚𝐥
External validation datasets are essential to assess true generalizability. Out-of-distribution (OOD) testing reveals robustness across different scanners, labs, and biological variations.
💡 𝐊𝐞𝐲 𝐓𝐚𝐤𝐞𝐚𝐰𝐚𝐲: While foundation models significantly reduce labeled data requirements, careful fine-tuning strategies and rigorous validation remain essential for ensuring generalizability in real-world pathology applications.
|
|
|
|
|
|
|