Hierarchical Task-Driven Feature Learning for Tumor Histology
Project Details
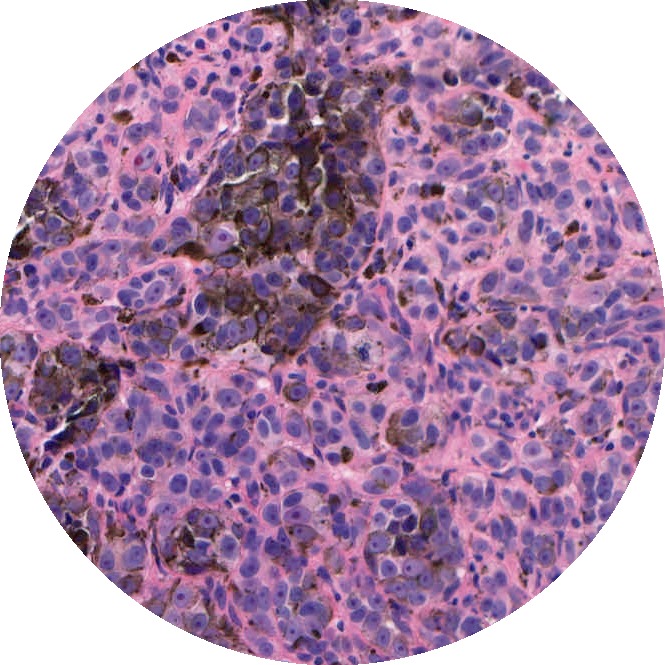
Background: Most automated analysis of histology follows a general pipeline of first segmenting nuclei, then characterizing color, texture, shape, and spatial arrangement properties of cells and nuclei. These hand-crafted features are time-consuming to develop and do not adapt easily to new data sets. More recent work has begun to learn appropriate features directly from image patches. This project was completed before deep learning tool kits were readily available and instead used dictionary learning as a means for learning representations from image patches.
Solution: Through learning small and large-scale image features, we captured the local and architectural structure of tumor tissue from histology images. This was done by learning a hierarchy of dictionaries, where each level captured progressively larger scale and more abstract properties. By optimizing the dictionaries further using class labels, discriminating properties of classes that are not easily visually distinguishable to pathologists were captured.
Results: We explored this hierarchical and task-driven model in classifying malignant melanoma and the genomic subtype of breast tumors from histology images. We also showed how interpreting our model through visualizations can provide insights to pathologists.
Additional Applications: Dictionary learning can be used as a discriminative representation for classification of textures, objects, scenes, or many other types of images. It can also form a compact descriptor for image retrieval. While not as powerful a discriminator as many deep learning methods, it is much more interpretable and so would be a suitable representation when insight into the features driving classification is important.