Multiple Instance Learning for Heterogeneous Images: Training a CNN for Histopathology
Project Details
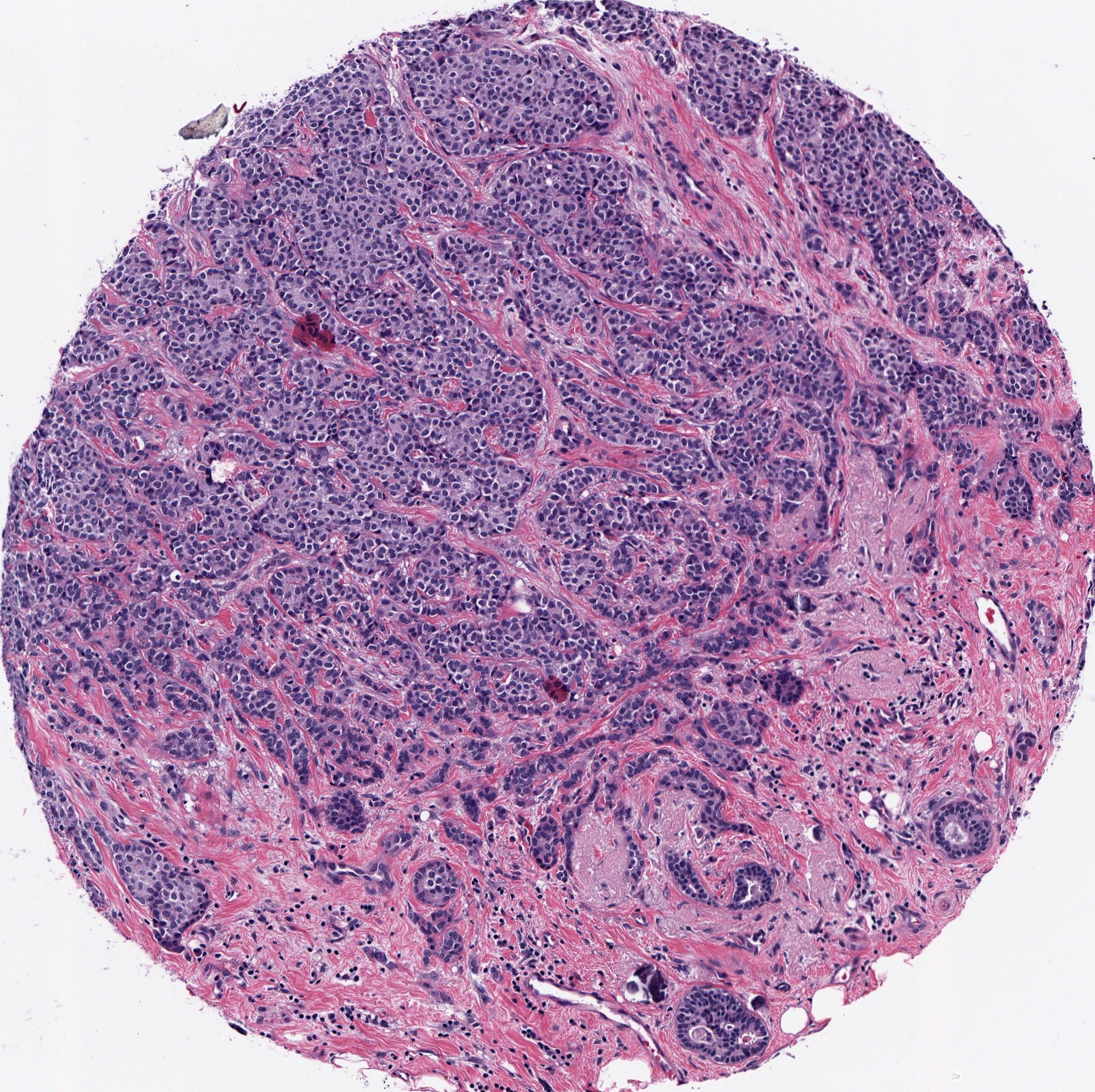
Background: Genomic subtypes for tumors are assigned from a single sample of tissue; they do not provide a spatial view of the tumor. Although some tumors are mostly homogeneous, others are very heterogeneous - both in appearance and molecular composition. Histology enables us to examine the spatial heterogeneity of the tumor in a way that gene expression cannot.
Challenge: Breaking large images into small regions for making class predictions is the first step in accommodating heterogeneous images. While prior work has explored different ways to aggregate predictions into a single image classification for some problem types, more general aggregation approaches are needed for heterogeneous images. Further, instance aggregation techniques in a deep learning setup can help adapt to this weakly labeled image scenario.
Solution: Multiple instance (MI) learning with a convolutional neural network enables end-to-end training in the presence of weak image-level labels. We developed a new method for aggregating predictions from smaller regions of the image into an image-level classification by using the quantile function. The quantile function provides a more complete description of the heterogeneity within each image, improving image-level classification.
Results: We validated our method on five different classification tasks for breast tumor histology and demonstrated a visualization method for interpreting local image classifications. Further, we examined predicted biomarker heterogeneity across the entire test set to show how our method may lead to future insights into tumor heterogeneity.
Additional Applications: Our methods can be applied whenever there are weak annotations for a concept due to the time or expense required for creating finer-grained annotations or due to the inability of humans to perform such an annotation task. More broadly, intra-class heterogeneity is present in many other classification problems from environmental to economic. Any time a label is applied to a group of cells, people, households, organisms, or other potentially diverse components, some amount of intra-class heterogeneity is present. The methods that we developed could provide insight into the amount of heterogeneity in a data set and, from that, a better understanding of the data itself.